MIT Biology Class - Reading Between the Lines (4)
Geneticists have been surprised to discover that some genes actually overlap each other, meaning that the same stretch of DNA can be involved in the code for more than one protein. This is very common in prokaryotes, which have less DNA in which to pack their genetic instructions, but it has also been discovered to occur in eukaryotes, which have a more roomy genome.
My thoughts:
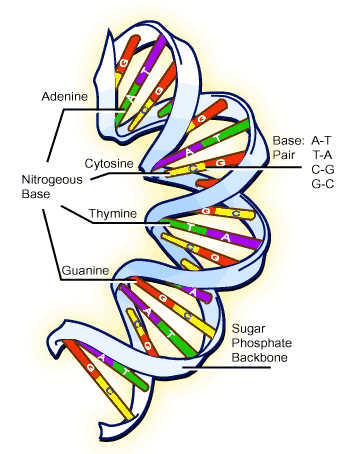
The amino acid arrangement, which makes up the protein polypeptide chain, is coded for by the nucleic acid molecules (nucleotides) in DNA. These nucleotides come in 4 flavors, which are represented by the letters A T G and C. A series of 3 nucleotides (a codon) codes for each individual amino acid. So, for example, TAC codes for tyrosine, while GAA codes for glutamate. It should be noted that there are 64 possible combinations of these sets of 3 nucleotides, while there are only 20 amino acids, so, many of them can be represented by more than one codon, e.g., both TAT and TAC code for tyrosine. Additionally, some combinations represent "start" and "stop" codons1, which bookend the gene and define a "reading frame" — the gene is contained within the reading frame.
A large molecule known as RNA polymerase reads the DNA and assists the messenger RNA in acquiring a copy of the gene, which will be elsewhere translated into a protein. It reads the DNA in these codon groups, but it is not as though it begins at the start of the DNA molecule and reads along it in groups of 3 nucleotides; it can attach at any point and only begins its transcription work when it encounters an ATG triplet (the start codon). So, in theory, there could be one at the start of the DNA strand, one starting from the 2nd nucleotide in the strand, one starting at the 3rd nucleotide in the strand, etc.
For example, a stretch of DNA might look something like this:
...ATGCATGTCATACCATAGCTAGAG...
The ATG at the beginning would start the gene sequence and it would end once it had reached a stop codon, which in this case is the TAG sequence. So the resulting reading frame and 4 amino acid gene2 would look like this:
...ATG CAT GTC ATA CCA TAG CTA GAG...
(Start, Histidine, Valine, Isoleucine, Proline, Stop)
But not so fast: if you look at the original sequence you will notice that ATG can be found a bit further in, and there is another TAG later down the line as well. So if we adjust our starting offset just a tad we can find a whole different gene in this same stretch of DNA, like so:
...A TGC ATG TCA TAC CAT AGC TAG AG...
(Start, Serine, Tyrosine, Histidine, Serine, Stop)
Can you see why this would be a stunner for geneticists to find? This is like discovering that your recipe for chicken pot pie also contained the recipe for laundry detergent if you drop every second letter, or that your favorite song is also the national anthem when played backwards. It is hard enough to explain how new genes can come into existence by random mutations; now we must explain how something so delicately intertwined as this could come about.
It is a problem whether it is claimed to come about gradually or all in one step. If you say that the overlapping genes came into existence together, then you have only increased the statistical improbabilities on already problematic equations. If you say that this happened gradually (first one gene appeared and then tweaks occurred to bring the second into play), then you have added profound constraints upon the process that is proposed to bring genes into existence. It is one thing to say that a functional gene could by chance and degree be constructed upon a stretch of unused DNA, but to say that it can arrive upon the back of a functioning gene without upsetting its vocation stretches credulity on principle alone. I may just as well say that my wife can slowly change her pot pie recipe into a crypto-detergent recipe without making my son vomit at any point that she follows it and served him the product.
~~~~~~~~~~~~
End Notes:
1. The start codon ATG actually codes for the amino acid methionine. The three stop codons, TAA, TAG, and TGA, do not encode an actual amino acid.
2. As you may remember from an earlier lecture note, the average protein consists of at least 150 amino acids.

22 Comments:
That is extraordinary. I would like to know how similar the overlapping genes are. I mean do they somehow compliment each other? Do they contribute to the same thing--say the length and width of a finger. Or are they as different as pot pie and laundry soap? I would think it would be far more extraordinary if they contribute to very different things, but that would just be an incredible coincidence that overlapping genes would both be functioning.
It's kind of like Bible Codes if you think about it. I mean if you read straight through the Bible, every word is meaningful. But if you start doing equa-distancing, it's only ever once in a while that you come across a word with meaning. And there are so many letters in the Bible, it's inevitable that you'd come across a meaningful word here and there. Bible Codes would be facinating only if equidistancing produced just as many meaningful words, sentences, and paragraphs as reading the Bible straight through. I think the same would apply to overlapping genes.
So, I have two questions. How common are overlapping genes? ...and... Do they contribute to similar functions or very different functions?
Thanks Paul,
That was fantastic!
More info here.
http://www.creationontheweb.com/content/view/5158/
Thanks, Duane, that is a worthy link, which discusses the further complexity and surprises beyond what I mention here. The trend in biological research is always toward uncovering more complexity. The obvious question to ask the evolutionist is, "Just how complex do we have to discover life to be before it becomes statistically impossible for random variation and selection, plus a finite amount of time, to be a viable explanation?" Some bailed on the discovery of DNA and its interdependency with RNA, ribosomes, and proteins. We have gone well beyond that by now.
Sam,
These overlapping genes are still being discovered and explored, so it is hard to answer this. I can say that I see evidence that they are being found to be much more common than thought, and one source even claimed they might turn out to be the rule rather than the exception. It seems also dependent upon the type of organism, but the prediction that it would be more abundant in compact genomes rather than more roomy genomes (like in humans) has not played out well. The hard numbers I found in some specific studies put it anywhere from .5% to 10%, depending upon the organism.
One reason that they first suspected this is that there seemed to be more proteins being produced than the number of genes that they had mapped — way more. But the thing that makes it hard to know for sure which extras are in the overlaps is that the known genes can actually be "spliced" in different ways (which is yet another problem of a similarly complex nature). Exactly how this splicing is driven is at the cutting edge of genetic science.
It should be noted that one stretch of DNA could theoretically contain 6 different reading frames: 3 forward and 3 backward! I haven't heard the extent of what they've found, but I do know they've found at least triple overlaps.
Another interesting fact that foils evolutionary predictions is this: because a single nucleotide change (a point mutation) is far more likely to impact something within overlapping gene-space, there should be fewer surviving neutral changes within overlapping genes than among non-overlapping ones (overlapping genes should be more highly "conserved"). This means that one would expect to see more differences in the non-overlapping genes between diverse creatures sharing certain genes than among the overlapping genes that they share. This is a logically sound prediction, but it has not proved to be true. The history of evolutionary theory is paved with such surprises and failed predictions.
Regarding whether or not these overlaps involve complementary functionality, I know very little. However, it has been found to be true in at least some cases. You may be interested in what are termed "operons", which are groups of functionally related genes that are consecutively positioned and which share regulatory processes. Since it is not uncommon for genes to be organized in these ways, it will not surprise me to find that overlaps show similar functional arrangement. An evolutionist would say that if it adds value, then selection would favor it, but it certainly does not make it any more probable for multiple related things to randomly appear together merely because it would be more beneficial if they did so.
The difference between this and the "Bible Codes" is that with equidistance text, you are finding things that just happen to be constructed from the mainline text, but are not detectable by the normal method of browsing that text (i.e., by reading it), and it is only arguably intended to be found in the text. However, with overlapping genes we are talking about multiple, fully-functional genes that are being read and utilized by the same machinery of the cell. Maybe it's more like discovering a tribe of people whose alphabet looks much like the English alphabet only upside down; and when one of them picks up a Bible, he turns it upside down and notices that a bunch of complete chapters are actually readable, theologically meaningful, and grammatically perfect.
Actually, there are 22 known amino acids -- meaning that the genetic code really is a code (its meaning depends upon context). That is, there is no chemical necessity that this particular codon codes for this particular amino acid. For, not only do multiple codons code for the same animo acid (as per the "Canonical," formerly "Universal," Genetic Code), but with the discoveries of the 21st and 22nd amino acids, it is seen that at least two codons code for more than one amino acid.
See, for example, this article, form 2002, on the discovery of the 22nd.
Thanks, Ilmon. Mention of this might have made another good footnote, i.e., that certain organisms and proteins utilize more than the "canonical" 20 amino acids in unique ways.
I'm not sure if you meant it as a footnote or as a problem for my conclusion. In my view, this only deepens the mystery. Exceptions to rules only mean further complexities that can impose themselves upon those rules. As this paper describes it, biosynthesis of selenocysteine (the 21st amino acid) requires "complex molecular machinery that recodes in-frame UGA codons, which normally function as stop signals." Pyrrolysine (the 22nd) is similarly synthesized in some organisms.
Not that it would make a difference to my case, but so far it does not appear to be true that "at least two codons code for more than one amino acid." These amino acids are coded for by two of the stop codons (rather, inserted at this point), not by codons which normally make another amino acid. But nothing will surprise me, and I will now go on record to predict that in 10 years everything that we now believe about protein expression and genetics will seem childishly simplistic. Though I will also predict that there is no level of complexity too great to overcome some people's faith in materialistic explanations.
Paul,
Whether we stretch our view into the infinite macro or magnify the unimaginably micro, complexity and wonder look back at us. Is there no place to find the illusive simpler that has to underlie the more complex? Everywhere, with every discovery we find that the fingerprints of an infinite mind already besmudge the pristine discovery.
This was the best, so far, of this series. Thanks for posting
Paul: "Mention of this might have made another good footnote, i.e., that certain organisms and proteins utilize more than the "canonical" 20 amino acids in unique ways."
Perhaps next time.
Paul: "I'm not sure if you meant it as a footnote or as a problem for my conclusion."
As more information pertinent to your conclusion ... after all, I'm a "wicked" (as per Dawkins) DarwinDenier.
Paul: "In my view, this only deepens the mystery. Exceptions to rules only mean further complexities that can impose themselves upon those rules."
Quite so. An exception to a rule means that the "rule" is incomplete (i.e. it isn't really a rule, but was mistaken to be one).
Paul: "Not that it would make a difference to my case, but so far it does not appear to be true that "at least two codons code for more than one amino acid." These amino acids are coded for by two of the stop codons (rather, inserted at this point), not by codons which normally make another amino acid."
Yes, I did mis-speak in saying "at least two codons code for more than one amino acid" (it was 2002 or 2003 when I last read one of the articles describing the 22nd amino acid; memory gets hazy, especially about facts with which one doesn't normally work). But, the point I was trying to bring to your attention is still correct: the meaning of a particular codon is context-dependent; there is no chemical necessity that *this* codon must code for *this* amino acid (or act as a "stop" signal) -- AND that this is already *known* to biologists.
Paul: "These amino acids are coded for by two of the stop codons (rather, inserted at this point), not by codons which normally make another amino acid."
However, I don't follow your parenthetical. The article I linked explicitly says:
"To qualify as genetically encoded, amino acids must be directly inserted into the growing peptide chain by a dedicated transfer RNA (tRNA) that recognizes a specific three-nucleotide codon in the messenger RNA (mRNA) transcript.
Pyrrolysine seems to qualify. ...
Smack in the middle of the MtmB gene--right where the pyrrolysine-specific codon is expected to be--is the stop codon UAG. But this particular UAG doesn't stop MtmB translation. Instead, a full-length protein is produced. ... "
Paul: "But nothing will surprise me, and I will now go on record to predict that in 10 years everything that we now believe about protein expression and genetics will seem childishly simplistic. Though I will also predict that there is no level of complexity too great to overcome some people's faith in materialistic explanations."
I'm confident that both your predictions will pan out.
For instance, at one time -- back when the genetic code was thought to be and was called "Universal," rather than "Canonical," as it's commonly called now -- some "Darwinists" asserted that the "fact" that the genetic code was "Universal" was *proof* that their materialist assumptions were correct. When further knowledge showed that the genetic code wasn't universal, after all, that claim was quietly forgotten.
(Just in case you haven't yet seen a reference to this) For another (recent) example of new knowledge punching holes in what we think we know about genetics and life processes -- it *appears* that inheritance isn't necessarily strictly genetic:
Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis: "... Here we show that Arabidopsis plants homozygous for recessive mutant alleles of the organ fusion gene HOTHEAD (HTH) can inherit allele-specific DNA sequence information that was not present in the chromosomal genome of their parents but was present in previous generations. ..."
Perhaps you'll enjoy some of the information presented on this page: The Genetic Codes
Also, I wonder about something related to "reading frames." You'd said to Sam: "It should be noted that one stretch of DNA could theoretically contain 6 different reading frames: 3 forward and 3 backward!"
But, I wonder if there might not potentially be 12 different "reading frames" on a given stretch of DNA -- a DNA molecule has two strands, after all (sometimes called "sense" and "non-sense" Designation of the two strands of DNA). Anyway, I vaguely recall (I won't insist this memory is correct) reading that biologists had identified that some regions on the "non-sense" strand are expressed.
Or, does the "backward" already refer to the "non-sense" strand? (In which case, my wonderment is answered)
Ah, nevermind that last (sorry if I'm gumming up the combox).
The mRNA is made from the "nonsense" strand (also called "antisense," also called "template") strand.
Ah (again). This page has tables that may better visually summarize the (known) non-canonical codes. Notice that while the majority of (known) non-canonical codes involve using one or more of the three canonical stop-codons as coding-codons, *not all* do.
Cenetic Codes
Ilion,
Thanks for that further info about the alternative coding. I believe I may actually have something in my class notes about this, but I can't easily review it because it is all captured in audio format. I think we've got two things going on here, though.
One is the insertion of the new amino acids on the occasion of one of the stop codons. As my referenced article stated it: "Translation of bacterial selenoprotein mRNA requires both a selenocysteine insertion sequence (SECIS) element, which is a stem-loop structure immediately downstream of Sec-encoding UGA codon, and trans-acting factors dedicated to Sec incorporation." In other words, it is not that UGA is translated by the conventional machinery as selenocysteine rather than as a stop codon; it is that there is additional machinery in the cell (e.g., SECIS) that intervenes to insert selenocysteine at this point.
The other related thing is that in some organisms it appears that the usual amino acid codons are consistently translated differently. The basic translation machinery would appear to be different. As you say, this was not an expected discovery by evolutionists. The reason is that an evolutionary change that converted one codon's translation into a different amino acid would have global effects upon the organism, which would certainly be lethal. For an analogy, imagine what would happen if we took any book in existence and converted all the "O"s to "A"s. The only way such a substitution would be at all helpful is if some of the words happened to be imperfectly spelled, like "cought" vs. "caught." However, it would have quite unfortunate effects upon words like "rope."
Regarding backward translation, it was indeed my understanding that when translation occurred in the opposite direction, it did so on the antisense strand. But I am feeling too lazy today to do my usual due-diligence to confirm that fact.
Paul, I presume that you have or will soon see this link at Uncommon Descent. But, just in case: Paramutation in mice
"A curious genetic phenomenon allows certain genetic instructions to be passed between generations without the gene variants involved being transmitted."
New Twist In MicroRNA Biology
"Professor Manolis Kellis and postdoctoral research fellow Alexander Stark report in the Jan. 1 issue of the journal Genes & Development that in certain DNA sequences, both strands of a DNA segment can perform useful functions, each encoding a distinct molecule that helps control cell functions. ..."
Folks in Hazard County ain't heard from Scott in a good long while.
Sorry Sam. I've been very negligent, haven't I? Between an extended illness, major vacation, work schedule, computer malfunctions, spring cleaning, and some rather addictive computer gaming with my son I've not been in blogger mode (or mood).
Interesting that you dropped a note, as I've been plotting my return of late. Here's hoping it's soon.
Excuses excuses. :-b
I'll be waitin'!
Forgive the intrusion, but I check regularly too. And would love to see a new blog entry. *cough, cough* You as well, Sam!
I've been missing you too! [sob]
Actually, Scott, your blog-neglect makes me feel not so bad for mine! :)
Post a Comment
<< Home